A research team led by Dr. Cao Xiaofeng at the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences has improved biomass-related traits in sheepgrass using its own custom genome editing system while increasing understanding of sheepgrass genomics.
The team’s genome assembly and innovative gene editing system, characterized by the fusion of big data and biotechnology, reveals the potential for intelligent and rapid genomic breeding of sheepgrass. The study was published in PNAS on Oct. 24.
Leymus chinensis (sheepgrass), a member of the Triticeae family, is a prominent grass species throughout the Eurasian steppe. This species, which is known for its robust rhizomes, has impressive attributes such as frost tolerance, drought tolerance, salt tolerance, and good soil stabilization capacity, etc.
Sheepgrass is widely recognized as a high quality forage that provides both ecological and economic benefits due to its exceptional nutritional value and palatability. However, due to the sheepgrass genome’s large size and high heterozygosity, studying it and elucidating its outstanding properties is challenging.
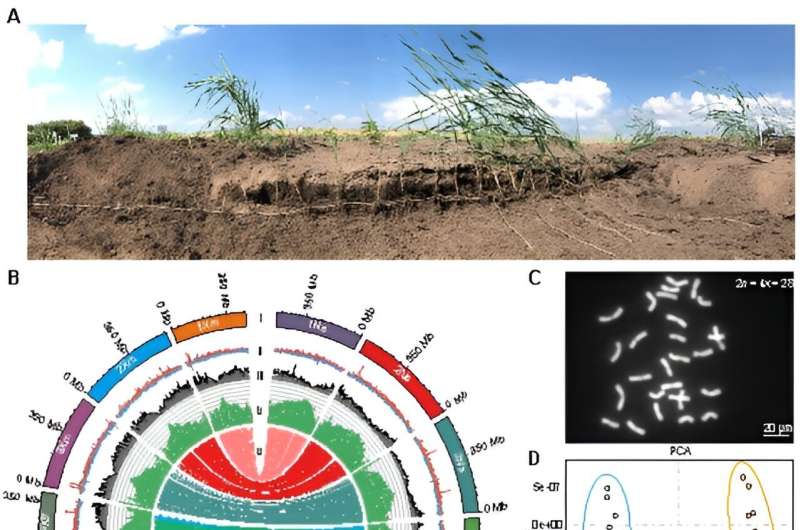
In this study, the researchers selected L. chinensis Lc6-5, a type of sheepgrass from the grasslands of Northeast China with strong rhizomes. They performed genome assembly using cutting-edge sequencing and assembly techniques. The assembled genome size was approximately eight Gb, with a contig N50 of more than 300 Mb and repetitive sequences accounting for 87.76% of the genome.
Given the remarkable heterozygosity of the genome, the researchers also performed a haplotype-level assembly. These results serve as an invaluable genomic resource for future research efforts.
Subsequently, the researchers used the custom CRISPR/Cas9 editing system on Lc6-5 to conduct a knockout of microRNA MIR528, which resulted in a significant increase in tiller number and plant growth rate.
The assembled sheepgrass genome, coupled with the genome editing system developed by the research team, opens the door to improving the biomass of L. chinensis and establishes a precedent for its rapid genetic improvement and for genome-based breeding.
More information:
Tong Li et al, Genome evolution and initial breeding of the Triticeae grass Leymus chinensis dominating the Eurasian Steppe, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2308984120
Citation:
Scientists use custom genome assembly and editing method to improve sheepgrass (2023, October 25)
retrieved 25 October 2023
from https://phys.org/news/2023-10-scientists-custom-genome-method-sheepgrass.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.

